
Quantitative in silico analytical chemistry, (Both in Japanese and English)
Quantitative Analysis of Molecular Interactions from Chromatography Retention to Enzyme Selectivity
Preface
Quantitatively explaining physicochemical phenomena is still a challenging subject in chemistry. We learn that wandering and understanding natural phenomena are a part of our permanent life. We started to use stone and wood as tools and modified them for easy handling. Then, we tried to imitate natural phenomena by ourselves. It was the time we learned to change to useful ones. Change is the beginning of chemistry. The synthesis of new compounds is the main target in chemistry. Then, we developed analytical instruments to understand the mechanisms of the changing process. We receive the analyzed results from a computer outlet of sophisticated instruments without their fundamental knowledge. To understand the phenomena quantitatively, we have to digitalize properties. Should we wait until someone prepares a textbook for the chemistry major? No, we should use presently available tools and up-to-date knowledge.
One available tool to obtain quantitatively molecular properties is computational chemistry, which specialists use to develop new drugs. However, computational chemists have not succeeded in creating new drugs even though they use very sophisticated computer systems. Fundamentally, drugs are usually inhibitors; therefore, we need pharmacological knowledge to recognize new molecules as drug candidates. One solution to this problem is exchanging the knowledge of many chemists who understand the chemical phenomena quantitatively. Therefore, education is the fundamental solution. We can build pyramids with many wide-knowledge people but not build super high buildings without specialists.
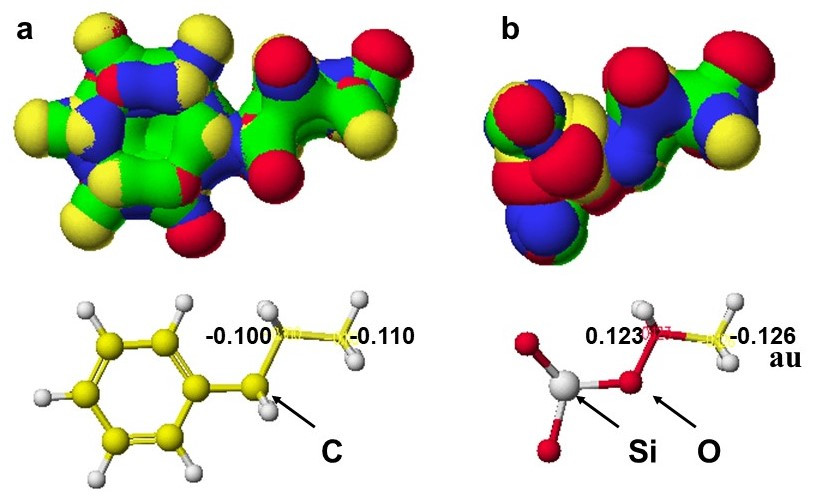
On the other hand, many students learn analytical chemistry, and bright students take creative chemistry courses for further study, where they can study to understand chemical phenomena quantitatively. The mysterious targets impress bright students. Analytical chemists should return to the starting point of understanding chemical phenomena quantitatively. Economical computational chemistry programs are available now. We should use them to learn and teach analytical chemistry, like organic chemistry. Organic chemists tried to explain the reactivity and summarized "The electronic theory of organic chemistry", and Hammett's s constants were derived from thin-layer chromatography data. Computational chemistry has upgraded the inductive effects. That is, the inductive effects are related to the chromatography retention difference. Why do analytical chemists not use computational chemistry to analyze the physico-chemical phenomena?
We predicted that computer-aided analytical chemistry was a popular tool 40 years ago, which proposed a computer controlling analytical instruments and handling analytical data. Developing analytical instruments have been dramatically improved, and automated intelligent analytical instruments provide high-precision results within a short time. We should use computers to analyze fundamental chemistry for advanced use of computers. The following chapters are a part of computational chemistry for learning analytical chemistry. Once students understand chemical phenomena written in analytical chemistry textbooks, they will create something new in the future. We live in a computer world, and we should use computers effectively for creation.
Chapter 1 explains the requirement of digitalizing molecular properties for quantitative explanations of analytical chemistry like organic chemists quantitatively explain organic reactions. The computational chemical analytical (in silico) method permits the digitalization of molecular properties, such as dipole-dipole, hydrogen bonding, Lewis acidity and basicity, dissociation, and enthalpy, to explain fundamental phenomena as the introduction study.
Chapter 2 quantitatively describes fundamental molecular interaction mechanisms using simple small molecules. The basic physical chemistry is "like dissolves like," and van der Waals force, electrostatic interaction, and hydrogen bonding are quantitatively explained as the calculated energy value differences using a computational chemistry program.
Chapter 3 describes the quantitative explanation of chromatographic retention, a tool for measuring molecular interactions with high reproducibility. Chromatography is a repeated phenomenon of adsorption and desorption. The mechanism can also be explained as a repeated partition. One side is solid or not movable liquid, and the other side is liquid or gas (carrier gas or solvent, eluent). The complicated mechanisms are quantitatively explained using simple model compounds, solvent molecules, and ions.
Chapter 4 exhibits the currently available computational chemical explanation of spectra analysis. Absorption spectra can be predicted using computational chemical calculations; however, these programs do not include solvents and buffers. Therefore, the feasibility is exhibited using color indicators. Furthermore, the application demonstrated for colorimetric analysis. NMR is improving the capability and determination of stereo structures of protein. Therefore, a combination of NMR spectroscopic analysis and computational chemical analysis becomes an attractive tool in protein chemistry. Molecular interaction is measured using NMR. However, the prediction of NMR spectra of small molecules is simple using the database computer program. Therefore, the feasibility of proton nuclear magnetic resonance (1H-NMR) and carbon nuclear magnetic resonance (13C-NMR) are described using relatively simple molecules for easy understanding.
However, some spectroscopic analytical methods are not included: Infrared spectra predicted using computational chemistry are too broad, and those predicted using database computer programs are too sharp; Mass spectra can be predicted using a database computer program without computational chemistry. Mechanisms of highly sensitive detections (chemiluminescence mechanisms and absorption detection of bromate in ion-chromatography and derivatization for highly sensitive detections) were described in the previous book.
Chapter 5 describes the preparation methods of model phases. 1 Quantitative analysis of chromatographic retention time requires model molecular interaction phases. The construction of stationary phases is complicated due to unknown structures of coated liquids that are generally a combination of solvents and ionic molecules. The thickness of the coated liquid depends on the surface material's properties and the mixture ratio of components. The model phases are prepared without adsorbed liquid. The details about gas chromatographic phases were described in the previous book.
Chapter 6 quantitatively describes the fundamental retention mechanisms of various liquid chromatography using model packing materials and solvent layers. Especially the competition between a model phase and a solvent layer for various molecules exhibited the actual mechanisms. The hydrophobicity, hydrogen bonding, and electrostatic are fundamental factors for the model phases, solvent layers, and analytes.
Chapter 7 quantitatively explains actual chromatographic data using molecular interaction energy values. The contribution of hydrogen bonding in the normal-phase, van der Waals force in the reversed-phase, and electrostatic in the ion-exchange liquid chromatography are clear from the difference of these molecular interaction energy values. The mechanisms in ion-pair liquid chromatography were a little complicated and required the combination.
The evaluation of column test methods to study the inactivation of chemically bonded silica gels is also described in detail.
The details of the Selectivity of the graphitized carbon phase, prediction, and chromatographic measurement of albumin-drug binding affinity were described in detail in the previous book. Affinity chromatography is not included in this chapter, but quantitative analysis of enzyme reactions is described in Chapter 9.
Chapter 8 describes new developments in chiral chromatography, including the enantiomer recognition mechanisms of polysaccharides, proteins, and natural organic chiral selectors. Designing and preparing chiral phases requires a wide range of chemistry knowledge; however, three-dimensional structures of chiral phases make clear the enantiomer recognition mechanisms. The enantiomer selectivity does not require high-plate number columns but requires a selective chiral phase and solvents based on the molecular recognition mechanisms. The details about Pirkle-type chiral selectors are described in the previous book.
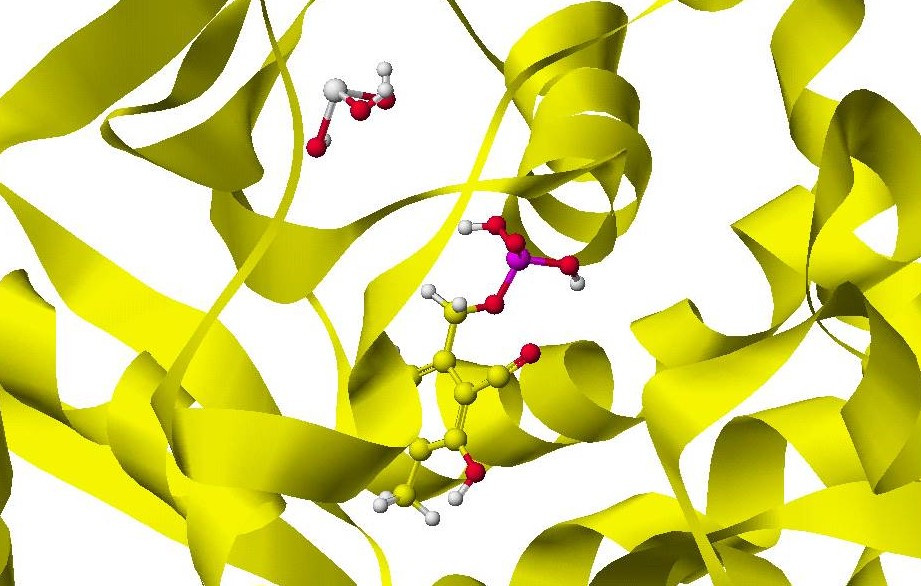
Chapter 9 quantitatively described the quantitative analysis of enzyme reactions as a part of the physical biochemistry of enzyme reactions. The basic phenomena of molecular recognition by proteins have been studied based on quantitative analysis of molecular interactions. Quantitative analysis of an enzyme's reactivity allows the design of mutants required to develop practical immunoassay methods, enzymatic biosensors, engineered enzymes, and new drugs. The reactivity is described based on electron localization, like in organic chemistry. Twelve enzyme reactivity was quantitatively described, including constructing human protein stereo structure from yeast protein structure and correcting and modifying PDB registered stereo structures.
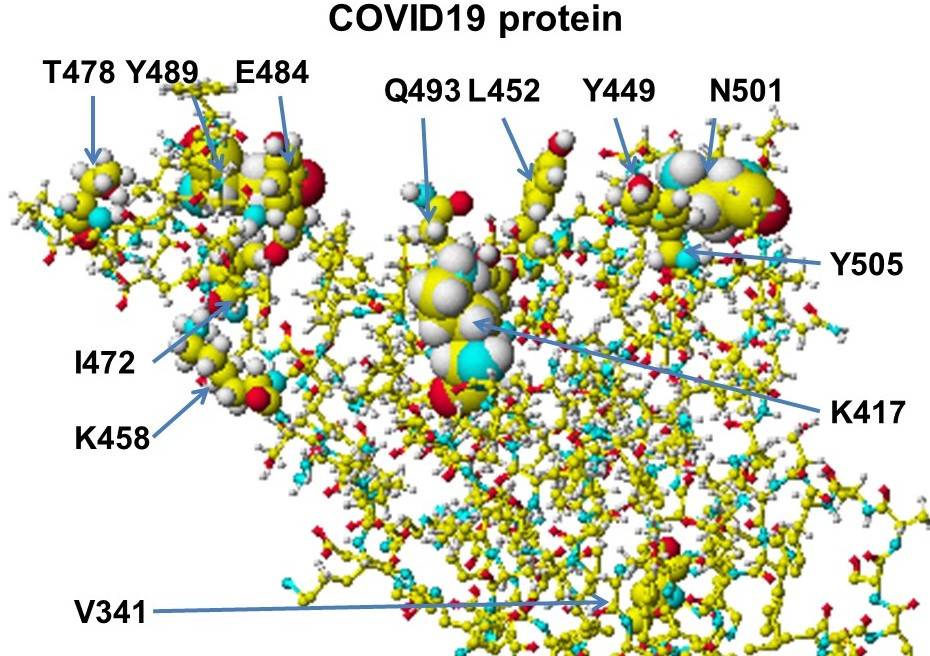
Chapter 10 exhibits an actual application of protein-protein interactions for the quantitative analysis of SARS-COVID-19 variants as the quantitative analysis of the transmissibility and the binding inhibitors. The binding strength (affinity) of variants was related to the strength of molecular interactions. Ion-ion interaction was the major contribution. Furthermore, various compounds were analyzed as binding inhibitors. The practical use of known medicines and the modification were described.
Total 327 pages including Index open online March 17, 2024 http://www.hanai-toshihiko.net
21 世紀 分析化学教室。Toward A.I. Analytical Chemistry
化学分析では多様な現象に基づく分析方法を開発して使用している。この分析方法の原 理を理解するためには定性的説明を定量的に説明する必要があり、分子構造に基づいて 計算化学で分子の特性を求めて、定量的(デジタル)に取り扱うことから始める。基本に なるのは分子の大きさ、ファンデルワールス体積で、これを基準にして置換基を付けれ ば分子の特性を考える手段となり、更に分子の特性が判ればより分析化学で扱う現象を 理解できる。有機化学の授業で以前から取り入れている方法である。
1. 滴定分析おける指示薬の色の変化の予測、比色分析用反応試薬の吸収スペクトル,
2. クロマトグラフィーに於ける保持の分子間相互作用の定量解析.
3. 医薬品開発で必要なアルブミンー薬物結合定数の予測,
4. 化学発光のように反応を伴う機構の解析,
5. 光学異性体選択性の定量解析、
6. 分子構造とスペクトル
7. 酵素反応の定量解析。
8. 蛋白質―蛋白質相互作用 COVID-19の感染力の定量解析と阻害剤の提案 オンラインで順次公開中
計算化学ソフトに含まれているプログラムを使って学習すれば幅広く化学を定量的に学習できるので入手しやすいプログラムを相談中です。水素結合を定量的に扱える計算化学 プログラムを使える人は練習問題を、無い人は読み飛ばしてください。
(公財)体質研究会 花井俊彦 http://www.hanai-toshihiko.net hanai104@kf7.so-net.ne.jp
- 2024 / 09 / 15 Covid-19 Transmissibility and the binding inhibitors
- 2024 / 09 / 15 Biomarkers and medicines
- 2024 / 08 / 24 Advanced HPLC
- 2024 / 03 / 17 Quantitative Analysis of Molecular Interactions from Chromatography Retention to Enzyme Selectivity
- 2021 / 11 / 15 計算化学を用いる分析化学 目次 1-19
- 2021 / 11 / 15 計算化学を用いる分析化学 20-37 HPLC, 光学異性体, 酵素反応, Covid19